Projects on biocide resistance in pests, weeds and vectors
Understanding adaptive evolution to herbicides in ryegrass
Intense pesticide and herbicide pressures have resulted in the evolution of resistance in multiples pests, insect vectors and weeds. Resistance is both a huge socio-economic issue limiting our capacity to control disease and sustain food production and the most obvious example of adaptive evolution, occuring within a human life-span. As resistance spreads, herbicides lose their efficacy and struggle to control weeds. This can reduce crop productivity and can represent a costly loss for farmers. Resistance is essentially a genetic response, it is best investigated at the genome level. Resistance is increasingly found to be endowed by multiple genes, which warrants to renew the set of tools available in weed research to incorporate recent advances in phenomics and genomics. We are aiming to identify all the mutations confering resistance using genome-wide association mapping and use these association to predict levels of resistance in any uncharacterised population of ryegrass based on genomic information

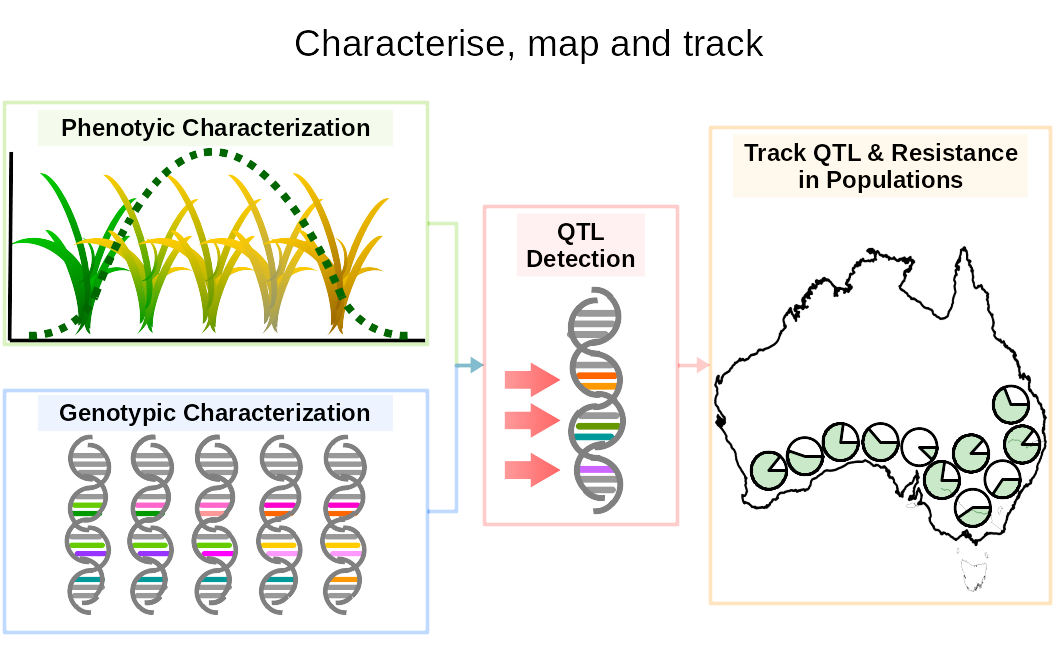
Exploring the potential of gene drives to control pest and invasive species
Australia has been and is vulnerable to biological invasion and resistance to control agents is a growing problem. Gene drives are emerging as powerful tools to transform the way we think population control. A gene drive is a selfish genetic element capable of self-replication and able to edit a specific allele and replace it with itself. Through this method, progeny of a gene drive individual will always inherit a copy of the gene drive, as will their progeny. After a number of generations, the gene drive may fixate. This could typically help to make currently ineffective herbicides useful again, by reintroducing a susceptibility allele in lieu of a resistance one.

To reintroduce susceptilibty, we need to be able to change the allele frequencies at the population scale. However, release of synthetic gene drives requires careful initial risk assessment. In collaboration with CSIRO, we contribute to building a risk assessment platform through the modelling of gene drive evolution using experimental genomic data. This way we can increase the specificity of the analysis to simulate case studies as close as possible to what would happen for the release of a gene drive in the real world.
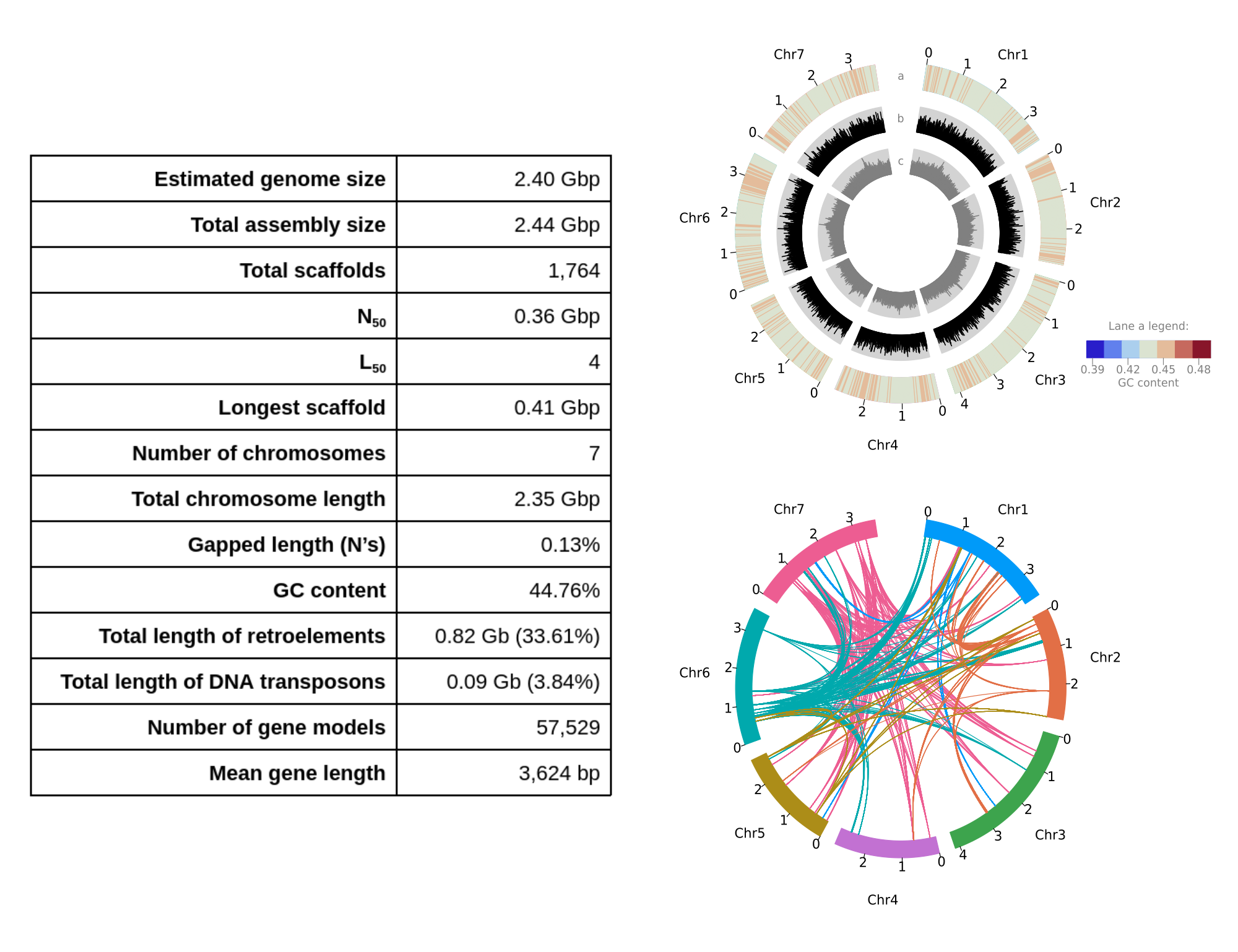
Lolium rigidum reference genome assembly
Download genome Genome browser
-
 Jeff Paril, Postdoctoral Associate
Jeff Paril, Postdoctoral Associate -
 Ben Camm, PhD candidate
Ben Camm, PhD candidate -
 Dr Jason Emms, GRDC
Dr Jason Emms, GRDC
People involved
Collaborators